Frontiers of Data and Computing ›› 2023, Vol. 5 ›› Issue (2): 37-49.
CSTR: 32002.14.jfdc.CN10-1649/TP.2023.02.003
doi: 10.11871/jfdc.issn.2096-742X.2023.02.003
• Special Issue: AI for Science • Previous Articles Next Articles
TU Youyou( ),ZHENG Qijing*(
),ZHENG Qijing*( ),ZHAO Jin*(
),ZHAO Jin*( )
)
Received:2023-02-15
Online:2023-04-20
Published:2023-04-24
Contact:
ZHENG Qijing,ZHAO Jin
E-mail:tyy2017@mail.ustc.edu.cn;zqj@ustc.edu.cn;zhaojin@ustc.edu.cn
TU Youyou,ZHENG Qijing,ZHAO Jin. Research on Quantum Proton Coupled Charge Transfer Process Based on Deep Neural Network[J]. Frontiers of Data and Computing, 2023, 5(2): 37-49, https://cstr.cn/32002.14.jfdc.CN10-1649/TP.2023.02.003.
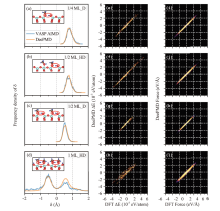
Fig.2
Comparison of DFT and DeePMD AIMD results. (a-d) Frequency density distributions of the proton transfer reaction coordinates δ in four adsorption structures. The molecular or interfacial sites where proton transfer may happen are circled in red in the sub-figures. The blue curves represent the frequency density distributions of δ in the original VASP AIMD datasets, and the orange curves represent the frequency density distributions of δ in the 50 ps AIMD trajectories calculated by DeePMD and i-Pi. (e-l) Comparison of DFT and DeePMD predicted energy (e-h) and force (i-l) of four structures on test sets. The energy has been shifted to the coordinate origin by subtracting the average value."
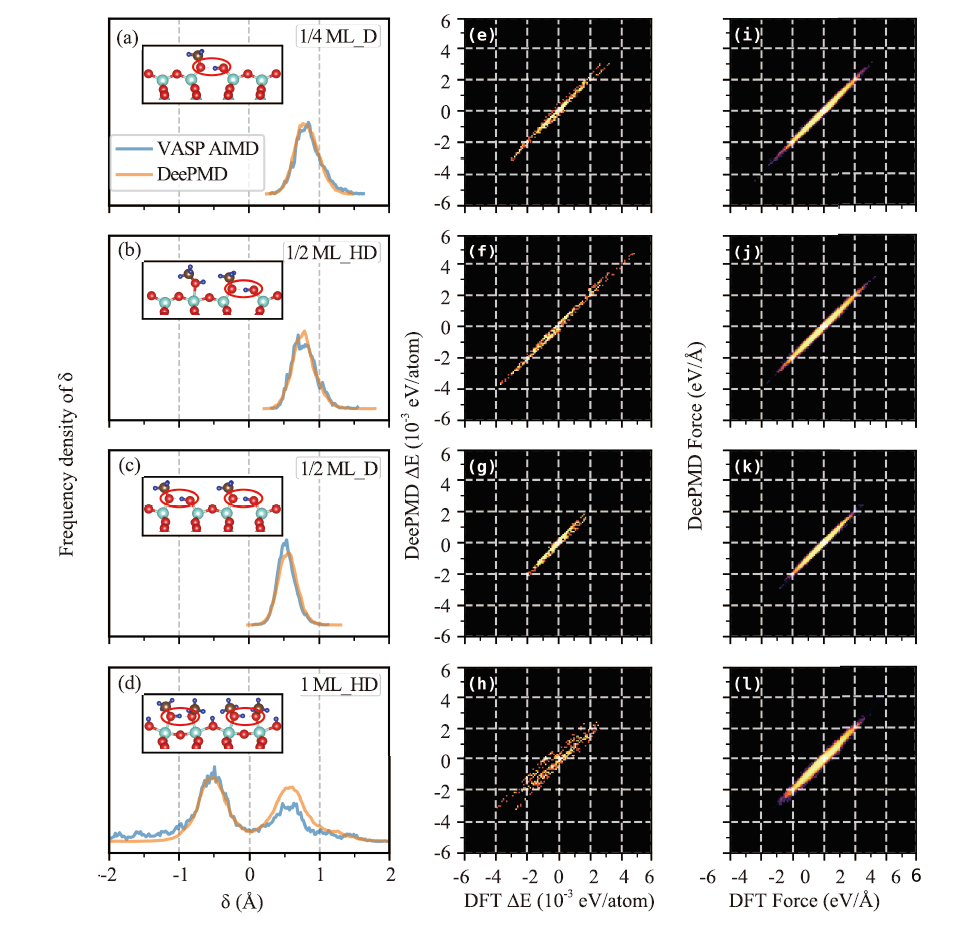
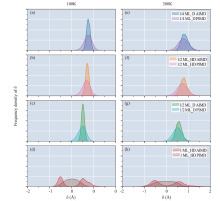
Fig.3
The 50 ps AIMD and 25 ps PIMD trajectories of four structures were obtained by combining the DeePMD training model with i-Pi. Frequency density distributions of the proton transfer reaction coordinates δ of the obtained four structures at (a-d) AIMD 100 K, (e-h) PIMD 100 K, (i-l) AIMD 300 K and (m-p) PIMD 300 K are shown."
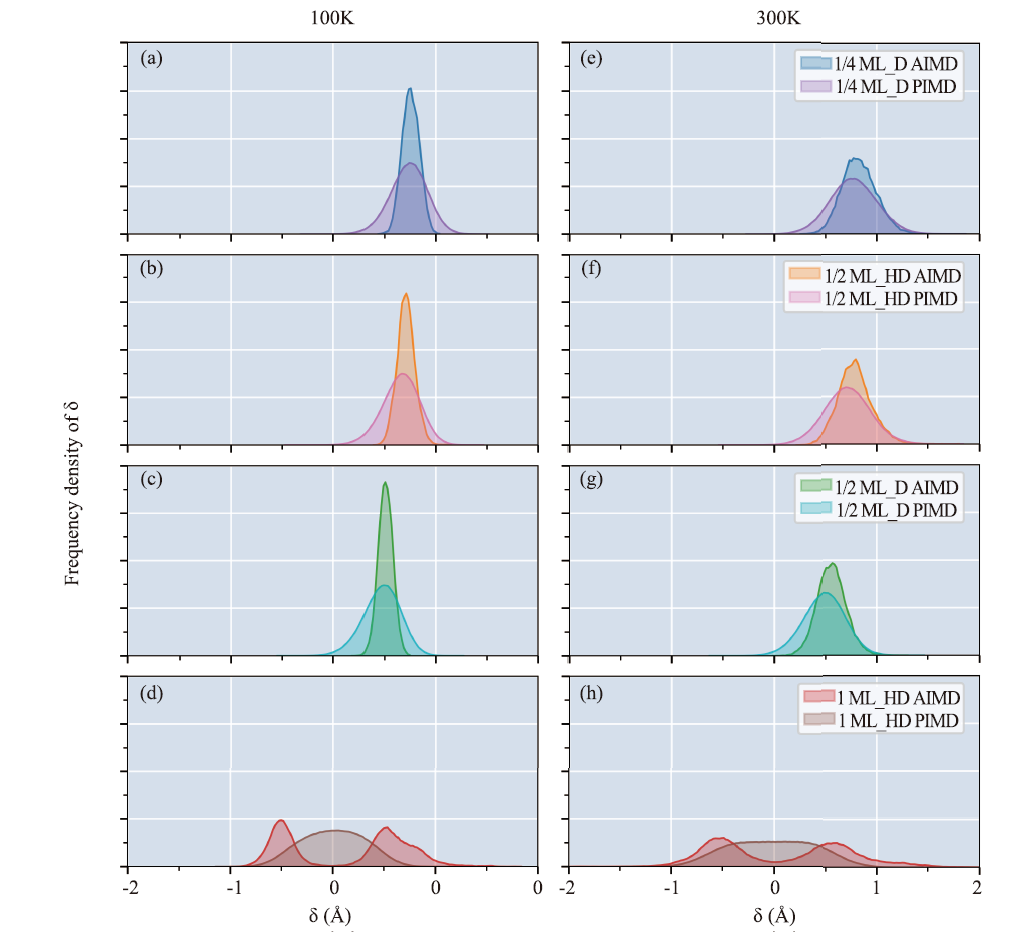
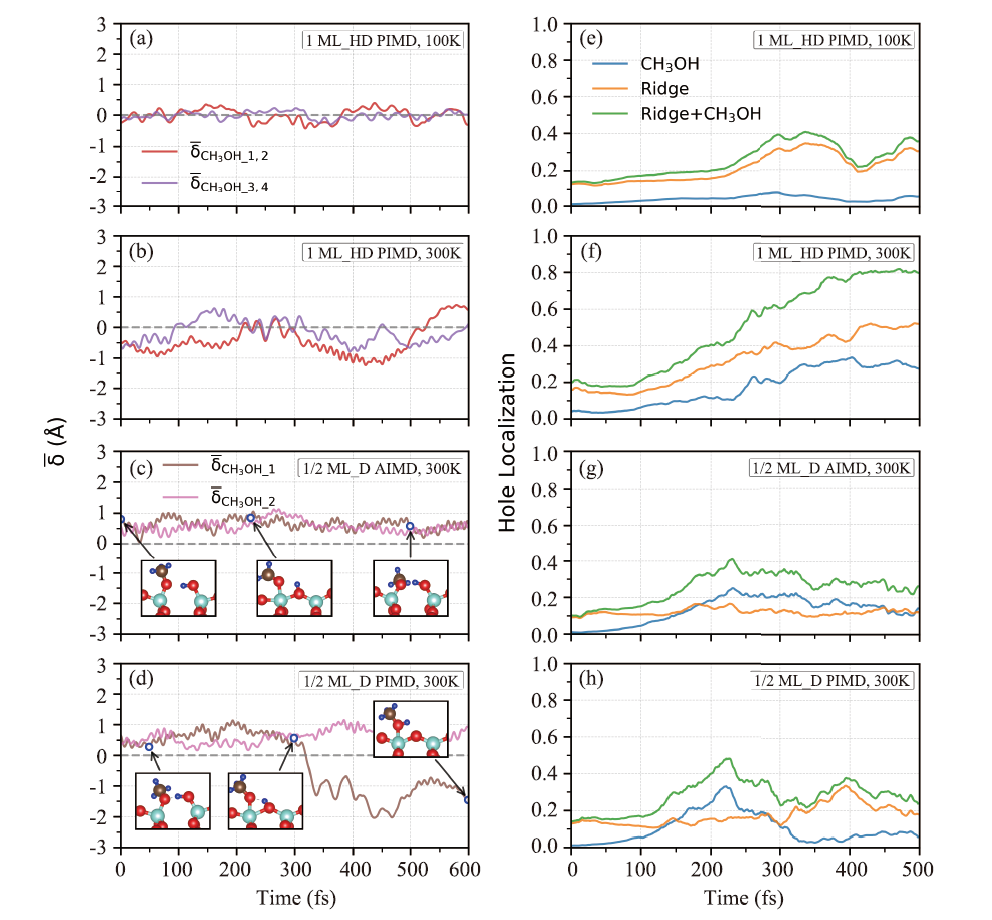
Fig.5
(a-b) Evolution of the centroid of the proton transfer reaction coordinates in the 1 ML_HD structure over 600 fs at 100 K and 300 K RPMD simulations. In RPMD is defined as the reaction coordinate of the average trajectory of each “bead” (i.e., the trajectory of the centroid of the “bead”). (c-d) Evolution of in the 1/2 ML_D structure over 600 fs at 300 K AIMD and RPMD simulations. (e-h) Hole transfer process corresponding to the molecular dynamics trajectories in (a-d), the evolution of holes in CH3OH. The ADM ridge structure and the overall distribution probability over time given by NAMD and RP-NAMD simulations are shown."
| [1] |
Petek H, Zhao J. Ultrafast Interfacial Proton-Coupled Electron Transfer[J]. Chemical Reviews, 2010, 110(12): 7082-7099.
doi: 10.1021/cr1001595 pmid: 21062076 |
| [2] | Zhao J, Yang J, Petek H. Theoretical study of the mo-lecular and electronic structure of methanol on a TiO2 (110) surface[J]. Physical Review B, 2009, 80(23): 235-416. |
| [3] |
Chen J, Li Y-F, Sit P, Selloni A. Chemical Dynamics of the First Proton-Coupled Electron Transfer of Water Oxidation on TiO2 Anatase[J]. Journal of the American Chemical Society, 2013, 135(50): 18774-18777.
doi: 10.1021/ja410685m pmid: 24308541 |
| [4] |
Karim W, Spreafico C, Kleibert A, Gobrecht J, Van-deVondele J, Ekinci Y, van Bokhoven J A. Catalyst sup-port effects on hydrogen spillover[J]. Nature, 2017, 541 (7635): 68-71.
doi: 10.1038/nature20782 |
| [5] |
Schrauben Joel N, Hayoun R, Valdez Carolyn N, Braten M, Fridley L, Mayer James M. Titanium and Zinc Oxide Nanoparticles Are Proton-Coupled Electron Transfer Age-nts[J]. Science, 2012, 336(6086): 1298-1301.
doi: 10.1126/science.1220234 pmid: 22679095 |
| [6] |
Li B, Zhao J, Onda K, Jordan K D, Yang J, Petek H. Ultrafast interfacial proton-coupled electron transfer[J]. Science, 2006, 311(5766): 1436-1440.
pmid: 16527974 |
| [7] |
Guerra W D, Odella E, Secor M, Goings J J, Urrutia M N, Wadsworth B L, Gervaldo M, Sereno L E, Moore T A, Moore G F, Hammes-Schiffer S, Moore A L. Role of Intact Hydrogen-Bond Networks in Multiproton-Coupled Electron Transfer[J]. Journal of the American Chemical Society, 2020, 142(52): 21842-21851.
doi: 10.1021/jacs.0c10474 pmid: 33337139 |
| [8] |
Wang L, Fried S D, Boxer S G, Markland T E. Quantum delocalization of protons in the hydrogen-bond network of an enzyme active site[J]. Proceedings of the National Academy of Sciences, 2014, 111(52): 18454-18459.
doi: 10.1073/pnas.1417923111 |
| [9] |
Schran C, Marsalek O, Markland T E. Unravelling the influence of quantum proton delocalization on electronic charge transfer through the hydrogen bond[J]. Chemical Physics Letters, 2017, 678: 289-295.
doi: 10.1016/j.cplett.2017.04.034 |
| [10] |
Guo J, Lü J T, Feng Y, Chen J, Peng J, Lin Z, Meng X, Wang Z, Li X Z, Wang E G, Jiang Y. Nuclear quantum effects of hydrogen bonds probed by tip-enhanced inel-astic electron tunneling[J]. Science, 2016, 352(6283): 321-325.
doi: 10.1126/science.aaf2042 |
| [11] |
Habershon S, Manolopoulos D E, Markland T E, Miller T F. Ring-Polymer Molecular Dynamics: Quantum Effects in Chemical Dynamics from Classical Trajectories in an Extended Phase Space[J]. Annual Review of Physical Chemistry, 2013, 64(1): 387-413.
doi: 10.1146/physchem.2013.64.issue-1 |
| [12] |
Cao J, Voth G A. The formulation of quantum statistical mechanics based on the Feynman path centroid density. IV. Algorithms for centroid molecular dynamics[J]. The Journal of Chemical Physics, 1994, 101(7): 6168-6183.
doi: 10.1063/1.468399 |
| [13] | Craig I R, Manolopoulos D E. Quantum statistics and classical mechanics: Real time correlation functions from ring polymer molecular dynamics[J]. The Journal of Che-mical Physics, 2004, 121(8): 3368-3373. |
| [14] |
Marx D, Parrinello M. Ab initio path integral molecular dynamics: Basic ideas[J]. The Journal of Chemical Physics, 1996, 104(11): 4077-4082.
doi: 10.1063/1.471221 |
| [15] |
Marx D, Parrinello M. Ab initio path-integral molecular dynamics[J]. Zeitschrift für Physik B Condensed Matter, 1994, 95(2): 143-144.
doi: 10.1007/BF01312185 |
| [16] |
Markland T E, Ceriotti M. Nuclear quantum effects enter the mainstream[J]. Nature Reviews Chemistry, 2018, 2(3): 0109.
doi: 10.1038/s41570-017-0109 |
| [17] |
Kresse G, Hafner J. Ab initio molecular dynamics for liquid metals[J]. Physical Review B, 1993, 47(1): 558-561.
doi: 10.1103/physrevb.47.558 pmid: 10004490 |
| [18] |
Kresse G, Hafner J. Ab initio molecular dynamics for open-shell transition metals[J]. Physical Review B, 1993, 48(17): 13115-13118.
doi: 10.1103/physrevb.48.13115 pmid: 10007687 |
| [19] |
Kresse G, Hafner J. Ab initio molecular-dynamics sim-ulation of the liquid-metal--amorphous-semiconductor transition in germanium[J]. Physical Review B, 1994, 49(20): 14251-14269.
doi: 10.1103/physrevb.49.14251 pmid: 10010505 |
| [20] |
Kresse G, Joubert D. From ultrasoft pseudopotentials to the projector augmented-wave method[J]. Physical Review B, 1999, 59(3): 1758-1775.
doi: 10.1103/PhysRevB.59.1758 |
| [21] |
Tu Y, Chu W, Shi Y, Zhu W, Zheng Q, Zhao J. High Photoreactivity on a Reconstructed Anatase TiO2 (001) Surface Predicted by Ab Initio Nonadiabatic Molecular Dynamics[J]. The Journal of Physical Chemistry Letters, 2022, 13(25): 5766-5775.
doi: 10.1021/acs.jpclett.2c01417 |
| [22] |
Zhang L, Han J, Wang H, Car R, E W. Deep Potential Molecular Dynamics: A Scalable Model with the Accu-racy of Quantum Mechanics[J]. Physical Review Letters, 2018, 120(14): 143001.
doi: 10.1103/PhysRevLett.120.143001 |
| [23] | Zhang L, Han J, Wang H, Saidi W, Car R. End-to-end symmetry preserving inter-atomic potential energy model for finite and extended systems[C]. Advances in Neural Information Processing Systems, 2018, 31. |
| [24] |
Wang H, Zhang L, Han J, E W. DeePMD-kit: A deep learning package for many-body potential energy repre-sentation and molecular dynamics[J]. Computer Physics Communications, 2018, 228: 178-184.
doi: 10.1016/j.cpc.2018.03.016 |
| [25] |
Ceriotti M, More J, Manolopoulos D E. i-PI: A Python interface for ab initio path integral molecular dynamics simulations[J]. Computer Physics Communications, 2014, 185(3): 1019-1026.
doi: 10.1016/j.cpc.2013.10.027 |
| [26] |
Kapil V, Rossi M, Marsalek O, Petraglia R, Litman Y, Spura T, Cheng B, Cuzzocrea A, Meißner R H, Wilkins D M, Helfrecht B A, Juda P, Bienvenue S P, Fang W, Kessler J, Poltavsky I, Vandenbrande S, Wieme J, Corm-inboeuf C, Kühne T D, Manolopoulos D E, Markland T E, Richardson J O, Tkatchenko A, Tribello G A, Van Speybroeck V, Ceriotti M. i-PI 2.0: A universal force engine for advanced molecular simulations[J]. Computer Physics Communications, 2019, 236: 214-223.
doi: 10.1016/j.cpc.2018.09.020 |
| [27] | Zheng Q, Chu W, Zhao C, Zhang L, Guo H, Wang Y, Jiang X, Zhao J. Ab initio nonadiabatic molecular dynamics investigations on the excited carriers in condensed matter systems[J]. Wiley Interdisciplinary Reviews: Compu-tational Molecular Science, 2019, 9(6): e1411. |
| [28] | Akimov A V, Prezhdo O V. The PYXAID Program for Non-Adiabatic Molecular Dynamics in Condensed Matter Systems[J]. Journal of Chemical Theory and Com-putation, 2013, 9(11): 4959-4972. |
| [29] | Akimov A V, Prezhdo O V. Advanced Capabilities of the PYXAID Program: Integration Schemes, Decoh-erence Effects, Multiexcitonic States, and Field-Matter Interaction[J]. Journal of Chemical Theory and Compu-tation, 2014, 10(2): 789-804. |
| [30] | Chu W, Tan S, Zheng Q, Fang W, Feng Y, Prezhdo O V, Wang B, Li X-Z, Zhao J. Ultrafast charge transfer coupled to quantum proton motion at molecule/metal oxide interface[J]. Science Advances, 2022, 8(24): eabo-2675. |
| [1] | LI Yan,HE Hongbo,WANG Runqiang. A Survey of Research on Microblog Popularity Prediction [J]. Frontiers of Data and Computing, 2023, 5(2): 119-135. |
| [2] | LIU Yunfan,LI Qi,SUN Zhenan,TAN Tieniu. Face Age Editing Methods Based on Generative Adversarial Network: A Survey [J]. Frontiers of Data and Computing, 2023, 5(2): 2-23. |
| [3] | XU Songyuan,LIU Feng. ESDRec: A Data Recommendation Model for Earth Big Data Platform [J]. Frontiers of Data and Computing, 2023, 5(1): 55-64. |
| [4] | CHEN Qiong,YANG Yong,HUANG Tianlin,FENG Yuan. A Survey on Few-Shot Image Semantic Segmentation [J]. Frontiers of Data and Computing, 2021, 3(6): 17-34. |
| [5] | PU Xiaorong,HUANG Jiaxin,LIU Junchi,SUN Jiayu,LUO Jixiang,ZHAO Yue,CHEN Kecheng,REN Yazhou. A Survey on Clinical Oriented CT Image Denoising [J]. Frontiers of Data and Computing, 2021, 3(6): 35-49. |
| [6] | HE Tao,WANG Guifang,MA Tingcan. Discovering Interdisciplinary Research Based on Word Embedding [J]. Frontiers of Data and Computing, 2021, 3(6): 50-59. |
| [7] | ZHANG Yining,HE Hongbo,WANG Runqiang. A Survey on Popular Digital Audio Prediction Techniques [J]. Frontiers of Data and Computing, 2021, 3(4): 81-92. |
| [8] | CHEN Zijian,LI Jun,YUE Zhaojuan,ZHAO Zefang. Hybrid Recommendation Model Based on Autoencoder and Attribute Information [J]. Frontiers of Data and Computing, 2021, 3(3): 148-155. |
| [9] | XIAO Jianping,LONG Chun,ZHAO Jing,WEI Jinxia,HU Anlei,DU Guanyao. A Survey on Network Intrusion Detection Based on Deep Learning [J]. Frontiers of Data and Computing, 2021, 3(3): 59-74. |
| [10] | LI Xu,LIAN Yifeng,ZHANG Haixia,HUANG kezhen. Key Technologies of Cyber Security Knowledge Graph [J]. Frontiers of Data and Computing, 2021, 3(3): 9-18. |
| [11] | ZHAO Weiyu,ZHANG Honghai,ZHONG Bo. A Deep Learning Based Method for Remote Sensing Image Parcel Segmentation [J]. Frontiers of Data and Computing, 2021, 3(2): 133-141. |
| [12] | SHEN Biao,CHEN Yang,YANG Chen,LIU Bowen. Computer Vision Detection and Analysis of Mesoscale Eddies in Marine Science [J]. Frontiers of Data and Computing, 2020, 2(6): 30-41. |
| [13] | Ren Huiying,Wang Jing,Wang Yangang. Turbulence Modeling Based on AutoML [J]. Frontiers of Data and Computing, 2020, 2(4): 121-131. |
| [14] | Zhang Shenglin,Lin Xiaofei,Sun Yongqian,Zhang Yuzhi,Pei Dan. Research on Unsupervised KPI Anomaly Detection Based on Deep Learning [J]. Frontiers of Data and Computing, 2020, 2(3): 87-100. |
| [15] | Chen Lei,Yuan Yuan. Image Recognition of Agricultural Diseases Based on Deep Transfer Learning [J]. Frontiers of Data and Computing, 2020, 2(2): 111-119. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
